2 minutes
Optuna (Hyperparameter search)
Optuna is a fantastic hyperparameter optimization framework that speeds up the process of hyperparameter tuning. Optuna enables efficient hyperparameter optimization by adopting state-of-the-art algorithms for sampling the hyperparameters space and pruning efficiently unpromising trials (link to original paper).
Here we will re-use the Wine Quality public dataset available on the UCI website (see post about Regression models) and run a Random Forest model which we will evaluate using Mean Squared Error (MSE).
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from sklearn.ensemble import RandomForestRegressor
import optuna
data = pd.read_csv("data/winequality-white.csv", sep=';')
x_train, x_test, y_train, y_test = train_test_split(data.iloc[:,:-1],
data.iloc[:,-1],
test_size=0.3,
random_state=123)
# Define an objective with parameters to try in your model
def objective(trial):
# Define parameters, it can be integer, float or categorical (refer to Optuna documentation for more details)
param_grid = {
"n_estimators": trial.suggest_int("n_trees", 50, 500, step=50),
"max_depth": trial.suggest_int("max_depth", 1, 20, step=1),
"min_samples_split": trial.suggest_int("min_samples_split", 2, 20, step=1),
"min_samples_leaf": trial.suggest_int("min_samples_leaf", 1, 20, step=1),
"max_features": trial.suggest_categorical("max_features", ["sqrt", "log2", None]),
}
cv = KFold(n_splits=5, shuffle=True, random_state=123)
cv_scores = np.zeros(5)
for i, (fold_idx, oof_idx) in enumerate(cv.split(x_train, y_train)):
x_fold, y_fold = x_train.iloc[fold_idx], y_train.iloc[fold_idx]
x_oof, y_oof = x_train.iloc[oof_idx], y_train.iloc[oof_idx]
model = RandomForestRegressor(**param_grid, random_state=123)
model.fit(x_fold, y_fold)
y_pred_oof = model.predict(x_oof)
cv_scores[i] = mean_squared_error(y_oof, y_pred_oof, squared=False)
return np.mean(cv_scores)
# Run study
n_trials = 50
study = optuna.create_study(direction="minimize") # specify whether you want to minimize or maximize the objective
study.optimize(objective, n_trials=n_trials)Once the hyperparameter search has been completed, we can find out the best parameters:
print(f"Best hyperparameters: {study.best_trial.params}")
print(f"Best MSE: {study.best_trial.value}")
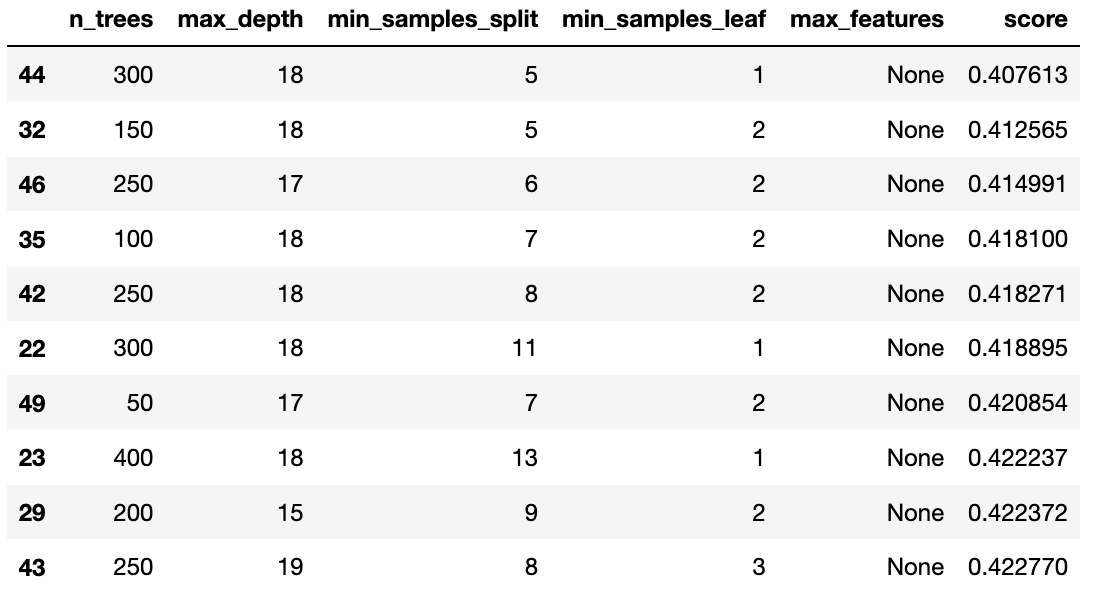
# Display the top 10 sets of parameters
res = pd.DataFrame([study.trials[i].params for i in range(n_trials)])
res['score'] = [study.trials[i].value for i in range(n_trials)]
res.sort_values('score').head(10)Best hyperparameters: {'n_trees': 300, 'max_depth': 18, 'min_samples_split': 5, 'min_samples_leaf': 1, 'max_features': None} Best MSE: 0.4076132896690571
And also get more visualization of the hyperparameter search:
optuna.visualization.plot_optimization_history(study)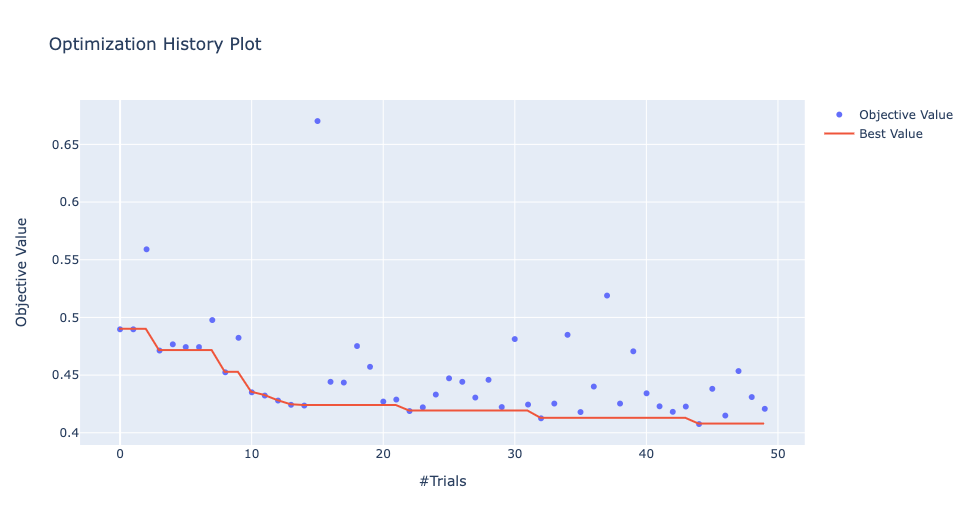
optuna.visualization.plot_slice(study)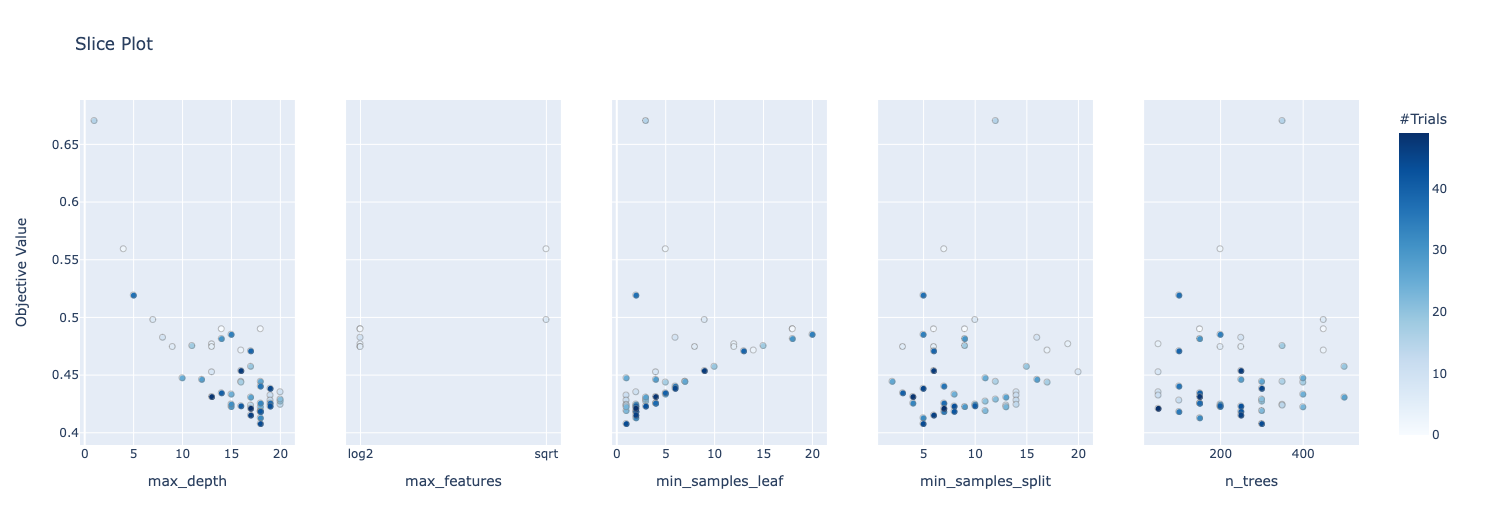
302 Words
2024-03-22 02:04